Contents
CRFB2_Stevens_fun.m
Examine the concern that the ARLs are systematically biased because the exponent of the Stevens function for motion and texture differs from 1.
File: work/MLExper/CRFeedback2/analysis/CRFB2_Stevens_fun.m
Usage: publish('CRFB2_Stevens_fun.m','html') ; % 'latex','doc','ppt'
Date: 2010-06-02% 1.0.0 2010-06-02 AP: Wrote it
Get started
cd(fullfile(CRFeedback2_pathstr,'analysis')) ; fprintf('\n\nCRFB2_Stevens_fun executed on %s.\n\n',datestr(now)) ; fprintf('cd %s\n',pwd) ; clear all ;
CRFB2_Stevens_fun executed on 02-Jun-2010 15:15:02. cd /Users/apetrov/a/r/w/work/MLExper/CRFeedback2/analysis
Load the data
H_filename = fullfile(CRFeedback2_pathstr,'data','H.mat') ; D_filename = fullfile(CRFeedback2_pathstr,'data','D.mat') ; %recalculatep = true ; recalculatep = false ; if (recalculatep || ~exist(H_filename,'file')) if (recalculatep || ~exist(D_filename,'file')) %- Write down repository revision number fprintf('!/usr/local/bin/svn info \n') ; !/usr/local/bin/svn info %- Concatenate the individual data files into one master ASCII file fprintf('!cat sbj*.dat > raw_data.dat \n') ; !cat sbj*.dat > raw_data.dat %- Import the ASCII data files to Matlab fprintf('\nD=CRFB2_import_data ...') ; g = fullfile(CRFeedback2_pathstr,'data','raw_data.dat') ; D = CRFB2_import_data(g,[],P) ; %- Save D for future use save(D_filename,'D') ; fprintf('\nsave %s \n',D_filename) ; else fprintf('load %s \n',D_filename) ; load(D_filename) ; end assert(exist('D','var')==1) ; %- Calculate the average response levels fprintf('\nH = CRFB2_data_by_sbj(D) ...\n') ; [H,P,ARL_params] = CRFB2_data_by_sbj(D,1,P) ; %- Save H for future use save(H_filename,'H') ; fprintf('\nsave %s \n\n',H_filename) ; else fprintf('load %s \n\n',H_filename) ; load(H_filename) ; end assert(exist('H','var')==1) ; N_trials = 350 ; % per stimulus type N_sbj = length(H) ;
load /Users/apetrov/a/r/w/work/MLExper/CRFeedback2/data/H.mat
Isolate the MOTION stimulus-response pairs
S_mot = zeros(N_trials,N_sbj) ; R_mot = zeros(size(S_mot)) ; for k=1:N_sbj S_mot(:,k) = H(k).MOTION_data(:,4) ; % categ == fdbk R_mot(:,k) = H(k).MOTION_data(:,7) ; % resp end xtab2(S_mot(:),R_mot(:))
| 1 2 3 4 5 6 7 | Total Percent
---------+-------------------------------------------+--------------
1 | 1014 605 289 106 61 8 3 | 2086 14.5
2 | 443 638 542 298 123 27 3 | 2074 14.5
3 | 121 379 552 581 317 99 13 | 2062 14.4
4 | 33 143 409 601 499 295 70 | 2050 14.3
5 | 8 60 175 418 607 481 289 | 2038 14.2
6 | 5 13 75 226 397 649 661 | 2026 14.1
7 | 3 5 19 76 233 517 1161 | 2014 14.0
---------+-------------------------------------------+--------------
Total | 1627 1843 2061 2306 2237 2076 2200 | 14350
Percent | 11.3 12.8 14.4 16.1 15.6 14.5 15.3 | 100.0
Isolate the TEXTURE stimulus-response pairs
S_txt = zeros(N_trials,N_sbj) ; R_txt = zeros(size(S_mot)) ; for k=1:N_sbj S_txt(:,k) = H(k).TEXTURE_data(:,4) ; % categ == fdbk R_txt(:,k) = H(k).TEXTURE_data(:,7) ; % resp end xtab2(S_txt(:),R_txt(:))
| 1 2 3 4 5 6 7 | Total Percent
---------+-------------------------------------------+--------------
1 | 1829 171 8 3 2 1 | 2014 14.0
2 | 635 1159 191 26 12 1 2 | 2026 14.1
3 | 162 662 905 243 52 11 3 | 2038 14.2
4 | 44 223 708 746 258 65 6 | 2050 14.3
5 | 13 60 260 595 790 300 44 | 2062 14.4
6 | 5 1 45 193 544 949 337 | 2074 14.5
7 | 3 2 4 34 104 360 1579 | 2086 14.5
---------+-------------------------------------------+--------------
Total | 2691 2278 2121 1840 1762 1686 1972 | 14350
Percent | 18.8 15.9 14.8 12.8 12.3 11.7 13.7 | 100.0
Stevens functions
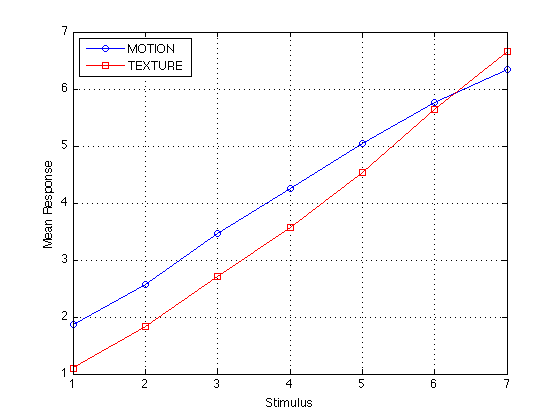
[s,S] = split(R_mot(:),S_mot(:)) ; mR_mot = aggreg(s) ; s = split(R_txt(:),S_txt(:)) ; mR_txt = aggreg(s) ; plot(S,mR_mot,'bo-',S,mR_txt,'rs-') ; grid on legend('MOTION','TEXTURE','location','NorthWest') ; xlabel('Stimulus') ; ylabel('Mean Response') ;
Correlations:
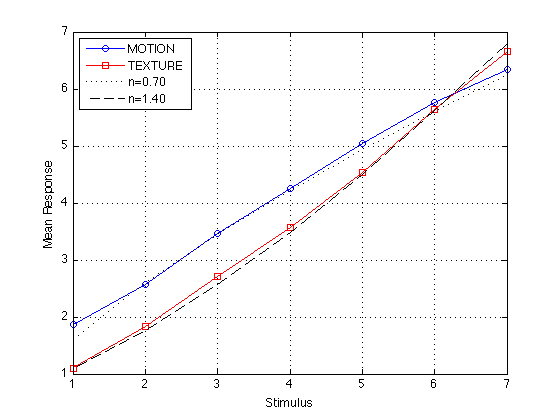
R070 = 1.6*S.^0.70 ; % n=0.70 -- arbitrarily chosen, not fit to data R140 = .7+(S.^1.4)./2.5 ; % n=1.40 -- arbitrarily chosen, not fit to data corrcoef([S mR_mot mR_txt R070 R140]) plot(S,mR_mot,'bo-',S,mR_txt,'rs-',S,R070,'k:',S,R140,'k--') ; grid on legend('MOTION','TEXTURE','n=0.70','n=1.40','location','NorthWest') ; xlabel('Stimulus') ; ylabel('Mean Response') ;
ans =
1.0000 0.9987 0.9980 0.9970 0.9956
0.9987 1.0000 0.9946 0.9982 0.9909
0.9980 0.9946 1.0000 0.9902 0.9994
0.9970 0.9982 0.9902 1.0000 0.9854
0.9956 0.9909 0.9994 0.9854 1.0000
Clean up
clear k s
Conclusion:
Even if the Stevens function is not linear, it is locally linear. The nonlinearity is negligible for the purposes of calculating the ARL. The correlation between the mean response (conditional on the stimuli) and the categories 1-7 is >.998 for both MOTION and TEXTURE.
% 2010-06-02