FINAL_ARL2_FITS.m
Figure 6 in .../papers/ProtoPaper/protopaper.tex Based on .../fdbk1/data/fit_ANCHOR_ARL2_part2.m and fit_INST_ARL2_part2.m Compare with .../fdbk1/data/model_ARL2_predictions.m
File: work/CMUExper/fdbk1/data/final_ARL2_fits.m Date: 2007-08-20 Alexander Petrov, http://alexpetrov.com
Contents
- Load empirical ARL2 profiles
- ANCHOR -- Explore 4 alternative sets
- INST
- Figure layouts
- Run 4 ANCHOR batches and 1 INST batches
- Summed Squared Error (SSE) and Root Mean SE (RMSE)
- Plot the group ARLs
- Define Assimilation := ARL(N)-ARL(P) and plot Assim profile
- Final Figure
- Clean up and archive workspace
Load empirical ARL2 profiles
clear all cd(fullfile(work_pathstr,'CMUExper','fdbk1','data')) ; load('S2.mat') ; ARL2 = [S2(:).ARL2] ; % 9x55 gr=[S2(:).group]' ; ugr = unique(gr) ; N_groups = length(ugr) ; ugr_descr = {'U1' 'L1' 'H1', 'L2' 'H2' } ; % [1 2 3, 5 6] fdbk_first_p = [1 1 1 , 0 0] ; % exp schedule by group %- Collapse across groups grARL2 = zeros(size(ARL2,1),N_groups) ; % 9x5 gr_idx = cell(1,N_groups) ; for k = 1:N_groups gr_idx{k} = find(gr==ugr(k))' ; grARL2(:,k) = mean(ARL2(:,gr_idx{k}),2) ; end empir_Assim = grARL2(:,[3 find(ugr==6)]) - grARL2(:,[2 find(ugr==5)]) ; N_runs_per_group = 250 ;
ANCHOR -- Explore 4 alternative sets
See ./fit_ANCHOR_ARL2_part2.m and ./html/fit_ANCHOR_ARL2_part2.html
H_set = [.030 .040 .050 .060] ;
mem_k_set = [.099 .083 .080 .065] ;
temper_set = [.022 .032 .046 .050] ;
aP = anchor_params ; % anchor model
N_cat = 7 ; aP.N_cat = N_cat ;
aP.anchors = default_anchors([.420 .660],N_cat) ;
aP.avail = [1:N_cat]' ;
aP.cutoffs = .60 .* [-3 -1 .75 3*.75] ;
aP.history = .040 ;
aP.mem_k = .083 ;
aP.temper = .032
aP =
scale: 'LINEAR'
N_cat: 7
SM_conv: 1.0000e-03
cat_sz: 0.0500
perc_k: 0.0400
mem_k: 0.0830
avail: [7x1 double]
anchors: [7x6 double]
cutoffs: [-1.8000 -0.6000 0.4500 1.3500]
temper: 0.0320
history: 0.0400
alpha: 0.3000
decay: 0.5000
ITI: 4
M_raster: 7
A_raster: [5 5 3 3]
mnfieldp: 1
INST
See ./fit_INST_ARL2_part2.m and ./html/fit_INST_ARL2_part2.html
iP = protoanchor_params([1:N_cat]) ; % INST model (aka "protoanchor")
iP.cutoffs = .60 .* [-3 -1 .70 3*.70] ;
iP.history = .050 ;
iP.mem_k = .050 ;
iP.temper = .030
iP =
scale: 'LINEAR'
N_cat: 7
SM_conv: 1.0000e-03
cat_sz: 0.0400
perc_k: 0.0400
mem_k: 0.0500
avail: [7x1 double]
instances: [7x3 double]
cutoffs: [-1.8000 -0.6000 0.4200 1.2600]
temper: 0.0300
history: 0.0500
alpha: 0.3000
decay: 0.5000
ITI: 4
Figure layouts
The figures below will have 6 subplots: 4 ANCHOR + 1 INST 3x2 layout: [a1 a3 i1 ; a2 a4 i2]
N_A_sets = length(H_set) ; % # of ANCHOR parameter sets
N_I_sets = 1 ;
Run 4 ANCHOR batches and 1 INST batches
fname = 'final_ARL2_fits.mat' ; if (file_exists(fname)) load(fname) ; else %- ANCHOR aSSE = zeros(1,N_A_sets) ; aRMSE = zeros(1,N_A_sets) ; for k = 1:N_A_sets H = H_set(k) ; mem_k = mem_k_set(k) ; temper = temper_set(k) ; aP.history = H ; aP.mem_k = mem_k ; aP.temper = temper ; [gr_aARL{k},aARL{k}] = run_ANCHOR_fdbk1(... 'model_params',aP,'groups',ugr','N_runs',N_runs_per_group) ; g = gr_aARL{k}(:,ugr) - grARL2 ; aSSE(k) = sum(g(:).^2) ; aRMSE(k) = sqrt(mean(g(:).^2)) ; end aSSE aRMSE %- INST iSSE = zeros(1,N_I_sets) ; iRMSE = zeros(1,N_A_sets) ; for k = 1:N_I_sets H = .050 ; mem_k = .050 ; temper = .030 ; iP.history = H ; iP.mem_k = mem_k ; iP.temper = temper ; [gr_iARL{k},iARL{k},Kargs] = run_INST_fdbk1(... 'model_params',iP,'groups',ugr','N_runs',N_runs_per_group) ; g = gr_iARL{k}(:,ugr) - grARL2 ; iSSE(k) = sum(g(:).^2) ; iRMSE(k) = sqrt(mean(g(:).^2)) ; end iSSE iRMSE ARL_params = Kargs.ARL_params ; end
Summed Squared Error (SSE) and Root Mean SE (RMSE)
aSSE = zeros(1,N_A_sets) ; aRMSE = zeros(1,N_A_sets) ; for k = 1:N_A_sets g = gr_aARL{k}(:,ugr) - grARL2 ; aSSE(k) = sum(g(:).^2) ; aRMSE(k) = sqrt(mean(g(:).^2)) ; end aSSE aRMSE iSSE = zeros(1,N_I_sets) ; iRMSE = zeros(1,N_I_sets) ; for k = 1:N_I_sets g = gr_iARL{k}(:,ugr) - grARL2 ; iSSE(k) = sum(g(:).^2) ; iRMSE(k) = sqrt(mean(g(:).^2)) ; end iSSE iRMSE
aSSE =
1.1265 1.0728 1.0693 1.1402
aRMSE =
0.1582 0.1544 0.1542 0.1592
iSSE =
1.3409
iRMSE =
0.1726
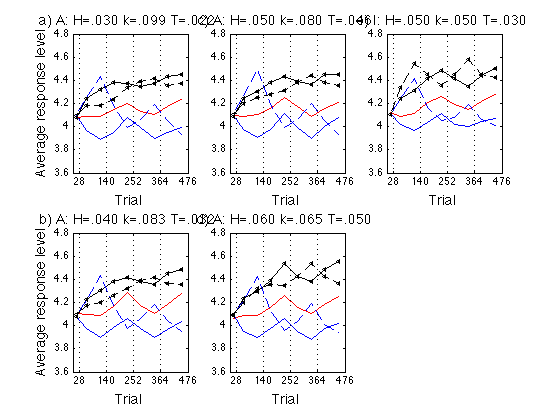
Plot the group ARLs
See .../fdbk1/data/model_ARL2_predictions.m
ls = {'r-' 'b-' 'k<-' 'r--' 'b--' 'k<--'} ; % group line styles
lw = [.5 1.5 1.5 .5 1.5 1.5] ; % group line widths
ANCHOR_subplot_idx = [1 4 2 5] ;
INST_subplot_idx = [3 6] ;
ANCHOR_panel = {'a)' 'b)' 'c)' 'd)'} ;
INST_panel = {'e)' 'f)'} ;
endpts = ARL_params.endpoints ; % [28 84 140 196 252 308 364 420 476]
step = endpts(3) - endpts(2) ;
x1 = endpts(1) ;
x = [x1/2 , [(endpts(2)+x1)/2:step:endpts(end)]]' ;
xtick = endpts(1:2:9) ;
%xtick = [0 endpts(1:2:9)] ;
ytick = [3.6:.2:4.8] ;
ax = [0 476 3.6 4.8] ;
clf ;
for k = 1:N_A_sets % ANCHOR
subpl = ANCHOR_subplot_idx(k) ;
subplot(2,3,subpl) ;
cla ; hold on ;
for g = ugr'
hh = plot(x,gr_aARL{k}(:,g),ls{g}) ;
set(hh,'LineWidth',lw(g),'MarkerSize',4) ;
end
hold off ; box on ;
hh = title(sprintf('%s A: H=.%03d k=.%03d T=.%03d ',...
ANCHOR_panel{k},round(H_set(k)*1000),...
round(mem_k_set(k)*1000),round(temper_set(k)*1000))) ;
set(hh,'FontSize',14) ;
axis(ax) ; set(gca,'xtick',xtick,'ytick',ytick,'xgrid','on') ;
if 1 % (subpl>=4)
hh = xlabel('Trial') ;
set(hh,'FontSize',14) ;
%else
% set(gca,'xticklabel',[]) ;
end
if (mod(subpl,3)==1)
hh = ylabel('Average response level') ;
set(hh,'FontSize',14) ;
%else
% set(gca,'yticklabel',[]) ;
end
end
for k = 1:N_I_sets % INST
subpl = INST_subplot_idx(k) ;
subplot(2,3,subpl) ;
cla ; hold on ;
for g = ugr'
hh = plot(x,gr_iARL{k}(:,g),ls{g}) ;
set(hh,'LineWidth',lw(g),'MarkerSize',4) ;
end
hold off ; box on ;
hh = title(sprintf('%s I: H=.%03d k=.%03d T=.%03d ',...
INST_panel{k},round(.050*1000),round(.050*1000),round(.030*1000))) ;
set(hh,'FontSize',14) ;
axis(ax) ; set(gca,'xtick',xtick,'ytick',ytick,'xgrid','on') ;
%set(gca,'yticklabel',[]) ;
if 1 % (subpl>=4)
hh = xlabel('Trial') ;
set(hh,'FontSize',14) ;
%else
% set(gca,'xticklabel',[]) ;
end
end
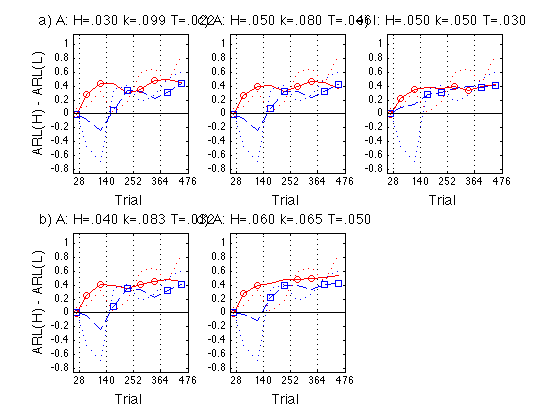
Define Assimilation := ARL(N)-ARL(P) and plot Assim profile
See .../fdbk1/data/model_ARL2_predictions.m
- 1 = Feedback first = Group 3 - Group 2
- 2 = No-feedback first = Group 6 - Group 5
ax = [0 476 -0.85 1.15] ; ytick1 = [-.8:.2:1] ; idx1 = [1 2 3 6 7] ; % feedback blocks for the feedback-first groups idx2 = [1 4 5 8 9] ; % feedback blocks for the no-feedback-first grps clf ; for k = 1:N_A_sets % ANCHOR Assim = gr_aARL{k}(:,[3 6]) - gr_aARL{k}(:,[2 5]) ; subpl = ANCHOR_subplot_idx(k) ; subplot(2,3,subpl) ; cla ; hold on ; hh = plot(x,Assim(:,1),'r-',x,Assim(:,2),'b--',... x(idx1),Assim(idx1,1),'ro',x(idx2),Assim(idx2,2),'bs',... x,empir_Assim(:,1),'r:',x,empir_Assim(:,2),'b:') ; set(hh(1:4),'LineWidth',1.5) ; axis(ax) ; hh=refline(0,0) ; set(hh,'Color','k','LineStyle','-','LineWidth',1) ; hold off ; box on ; hh = title(sprintf('%s A: H=.%03d k=.%03d T=.%03d ',... ANCHOR_panel{k},round(H_set(k)*1000),... round(mem_k_set(k)*1000),round(temper_set(k)*1000))) ; set(hh,'FontSize',14) ; axis(ax) ; set(gca,'xtick',xtick,'ytick',ytick1,'xgrid','on') ; if 1 % (subpl>=4) hh = xlabel('Trial') ; set(hh,'FontSize',14) ; %else % set(gca,'xticklabel',[]) ; end if (mod(subpl,3)==1) hh = ylabel('ARL(H) - ARL(L)') ; set(hh,'FontSize',14) ; %else % set(gca,'yticklabel',[]) ; end %hh = legend('Feedback first','No-feedback first',4) ; %set(hh,'FontSize',14) ; end for k = 1:N_I_sets % INST Assim = gr_iARL{k}(:,[3 6]) - gr_iARL{k}(:,[2 5]) ; subpl = INST_subplot_idx(k) ; subplot(2,3,subpl) ; cla ; hold on ; hh = plot(x,Assim(:,1),'r-',x,Assim(:,2),'b--',... x(idx1),Assim(idx1,1),'ro',x(idx2),Assim(idx2,2),'bs',... x,empir_Assim(:,1),'r:',x,empir_Assim(:,2),'b:') ; set(hh(1:4),'LineWidth',1.5) ; axis(ax) ; hh=refline(0,0) ; set(hh,'Color','k','LineStyle','-','LineWidth',1) ; hold off ; box on ; hh = title(sprintf('%s I: H=.%03d k=.%03d T=.%03d ',... INST_panel{k},round(.050*1000),round(.050*1000),round(.030*1000))) ; set(hh,'FontSize',14) ; axis(ax) ; set(gca,'xtick',xtick,'ytick',ytick1,'xgrid','on') ; if 1 % (subpl>=4) hh = xlabel('Trial') ; set(hh,'FontSize',14) ; %else % set(gca,'xticklabel',[]) ; end if (mod(subpl,3)==1) hh = ylabel('ARL(H) - ARL(L)') ; set(hh,'FontSize',14) ; %else % set(gca,'yticklabel',[]) ; end %hh = legend('Feedback first','No-feedback first',4) ; %set(hh,'FontSize',14) ; end
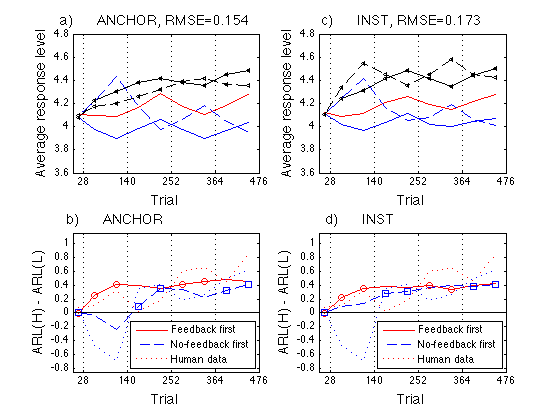
Final Figure
Single out ANCHOR parameter set #2 -- H=.040, mem_k=.083, T=.32
k = 2 ;
aP.history = H_set(k) ;
aP.mem_k = mem_k_set(k) ;
aP.temper = temper_set(k)
aAssim = gr_aARL{k}(:,[3 6]) - gr_aARL{k}(:,[2 5]) ;
iAssim = gr_iARL{1}(:,[3 6]) - gr_iARL{1}(:,[2 5]) ;
clf ;
ax = [0 476 3.6 4.8] ;
subplot(2,2,1) ; % ANCHOR ARL2
cla ; hold on ;
for g = ugr'
hh = plot(x,gr_aARL{k}(:,g),ls{g}) ;
set(hh,'LineWidth',lw(g),'MarkerSize',4) ;
end
hold off ; box on ;
hh = title(sprintf('a) ANCHOR, RMSE=%.3f ',aRMSE(k))) ;
set(hh,'FontSize',14) ;
axis(ax) ; set(gca,'xtick',xtick,'ytick',ytick,'xgrid','on') ;
hh = xlabel('Trial') ; set(hh,'FontSize',14) ;
hh = ylabel('Average response level') ; set(hh,'FontSize',14) ;
%hh = legend('U1','L1','H1','L2','H2',3) ; set(hh,'FontSize',14) ;
subplot(2,2,2) ; % INST ARL2
cla ; hold on ;
for g = ugr'
hh = plot(x,gr_iARL{1}(:,g),ls{g}) ;
set(hh,'LineWidth',lw(g),'MarkerSize',4) ;
end
hold off ; box on ;
hh = title(sprintf('c) INST, RMSE=%.3f ',iRMSE)) ;
set(hh,'FontSize',14) ;
axis(ax) ; set(gca,'xtick',xtick,'ytick',ytick,'xgrid','on') ;
hh = xlabel('Trial') ; set(hh,'FontSize',14) ;
hh = ylabel('Average response level') ; set(hh,'FontSize',14) ;
%hh = legend('U1','L1','H1','L2','H2',3) ; set(hh,'FontSize',14) ;
ax = [0 476 -0.85 1.15] ;
subplot(2,2,3) ; % ANCHOR Assim
cla ; hold on ;
hh = plot(x,aAssim(:,1),'r-',x,aAssim(:,2),'b--',...
x,empir_Assim(:,1),'r:',x,empir_Assim(:,2),'b:',...
x(idx1),aAssim(idx1,1),'ro',x(idx2),aAssim(idx2,2),'bs') ;
set(hh,'LineWidth',1.5) ;
axis(ax) ;
hh=refline(0,0) ; set(hh,'Color','k','LineStyle','-','LineWidth',1) ;
hold off ; box on ;
hh = title('b) ANCHOR ') ;
set(hh,'FontSize',14) ;
axis(ax) ; set(gca,'xtick',xtick,'ytick',ytick1,'xgrid','on') ;
hh = xlabel('Trial') ; set(hh,'FontSize',14) ;
hh = ylabel('ARL(H) - ARL(L)') ; set(hh,'FontSize',14) ;
hh = legend('Feedback first','No-feedback first','Human data',4) ;
%set(hh,'FontSize',14) ;
subplot(2,2,4) ; % INST Assim
cla ; hold on ;
hh = plot(x,iAssim(:,1),'r-',x,iAssim(:,2),'b--',...
x,empir_Assim(:,1),'r:',x,empir_Assim(:,2),'b:',...
x(idx1),iAssim(idx1,1),'ro',x(idx2),iAssim(idx2,2),'bs') ;
set(hh,'LineWidth',1.5) ;
axis(ax) ;
hh=refline(0,0) ; set(hh,'Color','k','LineStyle','-','LineWidth',1) ;
hold off ; box on ;
hh = title('d) INST ') ;
set(hh,'FontSize',14) ;
axis(ax) ; set(gca,'xtick',xtick,'ytick',ytick1,'xgrid','on') ;
hh = xlabel('Trial') ; set(hh,'FontSize',14) ;
hh = ylabel('ARL(H) - ARL(L)') ; set(hh,'FontSize',14) ;
hh = legend('Feedback first','No-feedback first','Human data',4) ;
%set(hh,'FontSize',14) ;
% Saved as work/papers/ProtoPaper/fig/final-fits.pdf
aP =
scale: 'LINEAR'
N_cat: 7
SM_conv: 1.0000e-03
cat_sz: 0.0500
perc_k: 0.0400
mem_k: 0.0830
avail: [7x1 double]
anchors: [7x6 double]
cutoffs: [-1.8000 -0.6000 0.4500 1.3500]
temper: 0.0320
history: 0.0400
alpha: 0.3000
decay: 0.5000
ITI: 4
M_raster: 7
A_raster: [5 5 3 3]
mnfieldp: 1
Clean up and archive workspace
clear hh k g Assim Kargs x1 ; clear aARL iARL ; % not used anyway save(fname) ;