fit_ANCHOR_ARL2_part2.m
Continued from fit_ANCHOR_ARL2_part1.m
Fit the ANCHOR2 model to the group ARL2 profiles of the Fdbk1 Experiment.
The main tool is .../work/models/anchor/fit_SSE/anchor_ARL2.m
File: work/CMUExper/fdbk1/data/fit_ANCHOR_ARL2_part2.m Date: 2007-08-20 Alexander Petrov, http://alexpetrov.com
Contents
Load the data
Start from scratch, generate new chev's, etc. The only thing used from fit_ANCHOR_ARL2_part1.m are the new c_ratio=.75 parameter setting and the new starting point for the searches.
clear all cd(fullfile(work_pathstr,'CMUExper','fdbk1','data')) ; load('S2.mat') ; ARL2 = [S2(:).ARL2] ; % 9x55 gr=[S2(:).group]' ; ugr = unique(gr) ; N_groups = length(ugr) ; ugr_descr = {'U1' 'L1' 'H1', 'L2' 'H2' } ; % [1 2 3, 5 6]
Collapse across groups
grARL2 = zeros(size(ARL2,1),N_groups) ; % 9x5 gr_idx = cell(1,N_groups) ; for k = 1:N_groups gr_idx{k} = find(gr==ugr(k))' ; grARL2(:,k) = mean(ARL2(:,gr_idx{k}),2) ; end
Fit INST on the same stimulus sequences, 11 per group
stim = [S2(:).dist] ; % 476x55
fdbk = [S2(:).fdbk] ;
Search parameters
Same search settings as in fit_ANCHOR_ARL2_part1.m Search strategy: 1. Fix c_ratio = .75 once and for all 1. Explore cutoff on a grid 2. Explore history on a grid 3. Search for mem_k and temper using FMINCON as before but start from mem_k = .085, temper = .030 % perc_k = .040 as always
fminconp = 1 ; flags = [0 1 1 0 0 0 0 0] ; % [perc_k mem_k temper hist cutoff xx xx c_ratio] Sparams = anchor_search_params2(flags,fminconp) ; optns = optimset(Sparams.optns,'TolX',1e-2,'Display','off') ; Sparams.optns = optns ; Sparams.target_vals = grARL2 c_ratio = .75 ; Sparams.params.cutoffs = .65 .* [-3 -1 c_ratio 3*c_ratio] ; Sparams.params.mem_k = .085 ; Sparams.params.temper = .030 ; Sparams.params.history = .040 ; Sparams.params
Sparams =
model_name: 'anchor_ARL2'
params: [1x1 struct]
p2v_templ: {'VAL = PARAMS.mem_k * 10 ;' 'VAL = PARAMS.temper * 10 ;'}
v2p_templ: {'PARAMS.mem_k = VAL / 10 ;' 'PARAMS.temper = VAL / 10 ;'}
bounds: [2x2 double]
ARL_params: [1x1 struct]
funfun_name: 'fmincon'
optns: [1x1 struct]
lsfun_name: 'sumsqerr'
target_vals: [9x5 double]
ans =
scale: 'LINEAR'
N_cat: 7
SM_conv: 1.0000e-03
cat_sz: 0.0500
perc_k: 0.0400
mem_k: 0.0850
avail: [7x1 double]
anchors: [7x6 double]
cutoffs: [-1.9500 -0.6500 0.4875 1.4625]
temper: 0.0300
history: 0.0400
alpha: 0.3000
decay: 0.5000
ITI: 4
M_raster: 7
A_raster: [5 5 3 3]
mnfieldp: 1
"Chance event" sets
N_chevs = 2 ; % Pack into "model data" structures suitable for passing as arguments. wfname = 'work_ANCHOR.mat' ; if (file_exists(wfname)) load(fname) ; else Mdata = cell(1,N_chevs) ; for k = 1:N_chevs Mdata{k}.stim = stim ; Mdata{k}.fdbk = fdbk ; Mdata{k}.group_idx = gr_idx ; %Mdata{k}.params = Sparams.params ; % will be set by SUMSQERR Mdata{k}.chev = make_anchor_chev(size(stim,1),size(stim,2),7) ; Mdata{k}.ARL_params = Sparams.ARL_params ; end end
Cutoff parameter grid
cutoff_levels = [.55 .60 .65 .70]' ; % c- in categ_sz units
N_cutoff_levels = length(cutoff_levels) ;
history_levels = [.020 .030 .040 .050 .060] ;
N_history_levels = length(history_levels) ;
Main work
fname = 'grid_search_ANCHOR_ARL2_part2.mat' ; if (file_exists(fname)) load(fname) ; else grid_SSE = zeros([N_cutoff_levels,N_history_levels,N_chevs]) ; grid_optX = cell([N_cutoff_levels,N_history_levels,N_chevs]) ; grid_det = cell([N_cutoff_levels,N_history_levels,N_chevs]) ; tic for k1 = 1:N_cutoff_levels c = cutoff_levels(k1) ; cutoffs = c .* [-3 -1 c_ratio 3*c_ratio] ; Sparams.params.cutoffs = cutoffs ; fprintf('\n cutoff %4.2f: ',c) ; for k2 = 1:N_history_levels H = history_levels(k2) ; Sparams.params.history = H ; for k = 1:N_chevs [par,SSE,optX,det] = paramsearch(Mdata{k},Sparams) ; % <-- Sic! grid_SSE(k1,k2,k) = SSE ; grid_optX{k1,k2,k} = optX ; grid_det{k1,k2,k} = det ; fprintf('%5.3f ',SSE) ; save(fname,'grid_SSE','grid_optX','grid_det','Sparams',... 'cutoff_levels','history_levels') ; end fprintf(', ') ; end end fprintf('\n') ; toc end % if file_exists ...
Plot the SSE
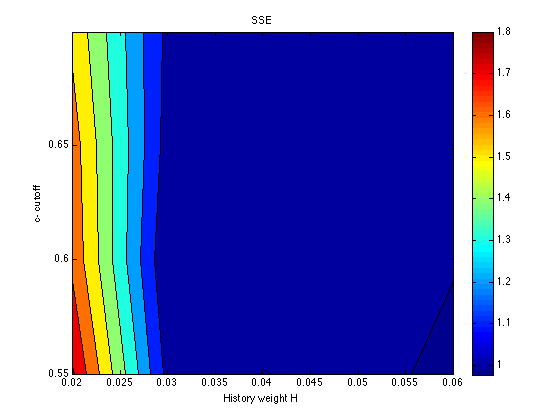
SSE = mean(grid_SSE,3) % average across the chev's [X,Y] = meshgrid(history_levels,cutoff_levels) ; contourf(X,Y,SSE) ; colorbar ; title('SSE') ; xlabel('History weight H') ; ylabel('c- cutoff') ;
SSE =
1.8085 1.0660 0.9972 1.0284 0.9776
1.6768 1.0070 1.0666 1.0177 1.0052
1.6504 1.0508 1.0680 1.0876 1.0769
1.5760 1.0721 1.0415 1.0580 1.0693
Plot the memory noise parameter
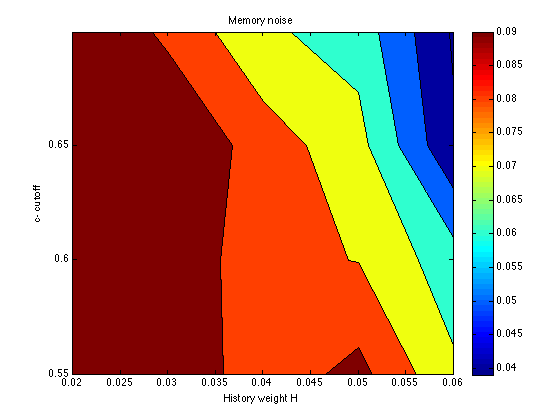
mem_k = zeros(size(grid_optX)) ; temper = zeros(size(grid_optX)) ; for k1 = 1:N_cutoff_levels for k2 = 1:N_history_levels for k = 1:N_chevs mem_k(k1,k2,k) = grid_optX{k1,k2,k}(1) / 10 ; temper(k1,k2,k) = grid_optX{k1,k2,k}(2) / 10 ; end end end mem_k = mean(mem_k,3) contourf(X,Y,mem_k) ; colorbar ; title('Memory noise') ; xlabel('History weight H') ; ylabel('c- cutoff') ;
mem_k =
0.1000 0.0984 0.0839 0.0932 0.0718
0.1000 0.0991 0.0827 0.0797 0.0646
0.1000 0.0997 0.0853 0.0738 0.0413
0.1000 0.0881 0.0717 0.0658 0.0390
Plot the temperature parameter
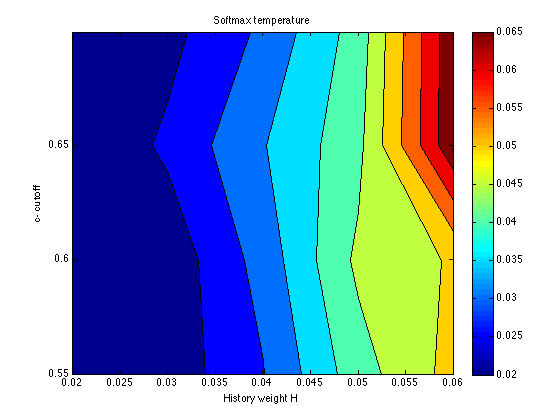
temper = mean(temper,3) contourf(X,Y,temper) ; colorbar ; title('Softmax temperature') ; xlabel('History weight H') ; ylabel('c- cutoff') ;
temper =
0.0200 0.0219 0.0297 0.0428 0.0517
0.0200 0.0217 0.0320 0.0461 0.0505
0.0200 0.0260 0.0347 0.0435 0.0687
0.0200 0.0234 0.0310 0.0421 0.0689
Conclusion
The SSE surface is quite flat in this region of the parameter space. The only region that is ruled out definitively is that for H<=.020. We are free to choose any history weight in the range .030-.060. This will be done to optimize (informally) the fit to the Assim profiles. We are also free to choose any cutoff in the range .55-.65. For roundness and specificity, let it be cutoff = .60. The other two parameters vary systematically as a function of H:
history: .030 .040 .050 .060 mem_k: .099 .083 .080 .065 temper: .022 .032 .046 .050
The fitting effort continues in ./final_ARL2_fits.m
Clean up
clear k k1 k2 X Y ;