Contents
- 010: Get started
- 015: Optimize parameters for figure?
- 020: Load H and sequence data
- 030: Create D structure
- 040: Create search_params structure
- 050: Optimize LRaven1_R2_goalfun
- 080: Run goalfun with optim params and then run global regression
- 090: Make quick and dirty plot of components, weights, and predictions
- 100: Generate publication-quality figures of components and weights
- 110: Generate and export high-quality systematicity component figure
- 120: Generate and export high quality toggle component figure
- 130: Generate and export high quality toggle weights figure
- 140:Generate and negative only weights
- 150: Generate and export high quality only positive weights figure
- 160: Generate and export high quality only negative weights figure
- 999: Finish
% LRAVEN1_comp_optim -- Use simplex to search for optimum R2/comp % % This is the script that generated Figures ?? and ?? in Hayes, Petrov, & % Sederberg (submitted). % % Script that is used to optimize the best 2 components in terms of R2. % It is meant to be used after a grid search has been performed to inform % the parameter boundaries. % % A simplex finds the best parameters, and then % generates publication quality component figures using export_fig. % % See also LRaven1_R2_optim, LRaven1_R2_publish, LRaven1_R2_goalfun, fminsearchcon % % (c) Laboratory for Cognitive Modeling and Computational Cognitive % Neuroscience at the Ohio State University, http://cogmod.osu.edu % % 1.1.0 2011-03-30 AAP: Fix lrate=.23 and gamma=22 -- see Section 015 % 1.0.1 2011-03-08 TRH: Clean up and finalize % 1.0.0 2011-01-26 TRH: Used as tool for pseudo-manual search for SR params
010: Get started
cd(fullfile(LRaven1_pathstr,'paramsearch')) ; fprintf('\n\nLRaven1_R2_publish executed on %s.\n\n',datestr(now)) ; fprintf('cd %s\n',pwd) ; clear all ; close all ;
LRaven1_R2_publish executed on 30-Mar-2011 19:35:21. cd /Users/apetrov/a/r/w/work/RelExper/LRaven1/matlab/paramsearch
015: Optimize parameters for figure?
Added 2011-03-30 by AAP.
When optimizep=false, the function will use the mean parameter values from the 35 cross-validation runs
%optimizep = true ; % call LRaven1_R2_paramsearch in Section 050 below optimizep = false ; % do no use the optimized params if (~optimizep) % This takes effect in Section 080 below [AAP 2011-03-30] % taken from ???? -- Taylor, please specify mean_CV_SR_params = [.22 0 .23] ; % [gamma lambda_gamma lrate] else mean_CV_SR_params = [NaN NaN NaN] ; % so that an error occurs if used below end
020: Load H and sequence data
% Load H: <1x35 struct> produced by LRaven1_hitcount filename = fullfile(LRaven1_pathstr,'data','H.mat') ; fprintf('load %s \n\n',filename) ; load(filename) ; clear filename % Load sequence data: <35x28 cell array> of transition sequence vectors trans_pathstr = fullfile(LRaven1_pathstr,'data','transitions') ; seq_filename = fullfile(trans_pathstr,'collapsed_clip_205.mat') ; %collapsed fprintf('load %s \n\n',seq_filename) ; load(seq_filename) ; clearvars trans_pathstr
load /Users/apetrov/a/r/w/work/RelExper/LRaven1/matlab/data/H.mat load /Users/apetrov/a/r/w/work/RelExper/LRaven1/matlab/data/transitions/collapsed_clip_205.mat
030: Create D structure
% Pack sequences into D D.sequences = recoded_sequences ; % Provide descriptor or sequences D.descriptor = ['Collapsed sequences 205 loaded from ' seq_filename] ; % Copy various possible score targets from H to D % These are not going to be used by LRaven1_R2_optim.m. Only D.target is % used. The rest are just stored along for the ride, and for post-hoc % analyses. D.session1_score = [H.session1_score]' ; D.session2_score = [H.session2_score]' ; D.session12_score = [H.session1_score]' + [H.session2_score]' ; % Single out which schores are TARGET during this particular run. D.target = D.session12_score ; % Print in the published transcript D %#ok<NOPTS> xtab1(D.target) % Size up the data -- SHOULD ALWAYS BE 35 x 28 [N_sbj,N_trials] = size(D.sequences) ;
D =
sequences: {35x28 cell}
descriptor: [1x125 char]
session1_score: [35x1 double]
session2_score: [35x1 double]
session12_score: [35x1 double]
target: [35x1 double]
Value Count Percent Cum_cnt Cum_pct
-------------------------------------------
12 1 2.86 1 2.86
13 1 2.86 2 5.71
15 1 2.86 3 8.57
16 1 2.86 4 11.43
18 1 2.86 5 14.29
19 2 5.71 7 20.00
20 3 8.57 10 28.57
21 4 11.43 14 40.00
22 3 8.57 17 48.57
23 2 5.71 19 54.29
24 7 20.00 26 74.29
25 3 8.57 29 82.86
26 5 14.29 34 97.14
27 1 2.86 35 100.00
-------------------------------------------
040: Create search_params structure
% AOI and component fields N_AOIs = 10 ; search_params.N_AOIs = N_AOIs ; % Number of unique interest areas N_comp = 2 ; search_params.N_comp = N_comp ; % Number of components % all sbjs search_params.max_N_comp = 12 ; % Maximum number of possible components % 12 sbj or less %search_params.max_N_comp = 6 ; % Maximum number of possible components % Subject and trial fields %search_params.sbj_idx = (1:2:N_sbj) ; % quick-and-dirty debugging search_params.sbj_idx = 1:N_sbj ; % Sbjs for analysis search_params.trial_idx = 1:28 ; % Trials for analysis % Search and Cross Validation fields search_params.N_left_out = 0 ; % Number of sbj left out for cross validation search_params.Borda_diagnosep = false ; % passed to select_components_borda.m fprintf('search_parasm.N_left_out=%d, which short-circuits the inner CV loop.\n', ... search_params.N_left_out ) ; search_params.N_random_seeds = 5 ; % Number of random param searches %search_params.N_random_seeds = 10 ; % quick-and-dirty debugging search_params.N_searches = 1 ; % Number of top seeds used for simplex run search_params.suggested_seeds = [.25 0 .23] ; % param start point(s) search_params.N_left_outermost = 1 ; % Cross valid. #sbj outer loop search_params.N_carryover_seeds = 0 ; % Number of seeds used in next iter. % Options and other fields search_params.verbosep = false ; % whether to display progress messages search_params.parallelp = false ; % whether to use parallel processing options = optimset('fmincon') ; % Set default options % Turn off display for parallel processing and set iterations to 50 search_params.options = optimset(options,'Display','iter','MaxIter',30) ; % File names fname_stem = 'R2opt-col-div-s12-Param-clamped-35x28-rigel-20110126' ; %fname_stem = 'delete-me' ; % use for debug search_params.log_streams = [1] ; %#ok<NBRAK> % only the console unless appended below search_params.log_title = ... 'Collapsed clip 205 sequences. All 35 Ss on all 28 trials. No inner CV loop.' ; % Set bounds on parameters search_params.gamma_bounds = [.01 .35] ; search_params.lambda_gamma_bounds = [0 0] ; search_params.lrate_bounds = [.01 .35] ; % Set LRaven1_R2_paramsearch 'diffp' field to false % This is the default behavior, but let's make it explicit that we intend % to use LRaven1_R2_goalfun inside LRaven1_R2_paramsearch. 2011-03-30 % % TO DO: Put a function handle to specify the goalfun explicitly. search_params.diffp = false ; % Display search_params in the published transcript search_params %#ok<NOPTS>
search_parasm.N_left_out=0, which short-circuits the inner CV loop.
search_params =
N_AOIs: 10
N_comp: 2
max_N_comp: 12
sbj_idx: [1x35 double]
trial_idx: [1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28]
N_left_out: 0
Borda_diagnosep: 0
N_random_seeds: 5
N_searches: 1
suggested_seeds: [0.2500 0 0.2300]
N_left_outermost: 1
N_carryover_seeds: 0
verbosep: 0
parallelp: 0
options: [1x1 struct]
log_streams: 1
log_title: 'Collapsed clip 205 sequences. All 35 Ss on all 28 trials. No inner CV loop.'
gamma_bounds: [0.0100 0.3500]
lambda_gamma_bounds: [0 0]
lrate_bounds: [0.0100 0.3500]
diffp: 0
050: Optimize LRaven1_R2_goalfun
% Optimize goalfun via LRaven1_R2_paramsearch. % This is not going to be used if optimizep=false, but it takes less than a % minute to generate and it's informative, so we calculate it ragardless. % It is going to be used when optimizep=true. [AAP 2011-03-30] tic , res = LRaven1_R2_paramsearch(D,search_params) , toc %#ok<NOPTS>
Iteration Func-count min f(x) Procedure
0 1 2.46029
1 3 2.46029 initial simplex
2 5 2.46029 contract inside
3 7 2.46029 contract inside
4 9 2.46029 contract inside
5 11 2.46029 contract inside
6 13 2.46029 contract outside
7 14 2.46029 reflect
8 16 2.45675 contract inside
9 18 2.45675 contract outside
10 20 2.45573 contract inside
11 21 2.45573 reflect
12 23 2.45561 contract inside
13 24 2.45561 reflect
14 26 2.45546 contract inside
15 28 2.45546 contract inside
16 30 2.45546 contract inside
17 32 2.45545 contract inside
18 34 2.45544 contract inside
19 36 2.45544 contract inside
20 38 2.45544 contract outside
21 40 2.45544 contract inside
22 42 2.45544 contract inside
23 44 2.45544 contract inside
24 46 2.45544 contract inside
25 48 2.45544 contract inside
26 50 2.45544 contract inside
27 52 2.45544 contract inside
28 54 2.45544 contract outside
29 56 2.45544 contract inside
30 58 2.45544 contract inside
Exiting: Maximum number of iterations has been exceeded
- increase MaxIter option.
Current function value: 2.455438
res =
search_params: [1x1 struct]
bounds: [2x3 double]
goalfun: @LRaven1_R2_goalfun
random_search_res: [5x7 double]
fminsearchcon_res: [1x1 struct]
exitflag: 0
top_cvRMSE: 2.4554
top_gamma: 0.2546
top_lambda_gamma: 0
top_lrate: 0.2329
opt_SR_params: [0.2546 0 0.2329]
Elapsed time is 45.584642 seconds.
080: Run goalfun with optim params and then run global regression
This is a repeat of the final call to LRaven1_R2_goalfun that ocurred inside LRaven1_R2_paramsearch. It is necessary to re-run the goalfun to get the export structure with the detailed components, etc.
% Commit to a parameter vector for the figures if (optimizep) SR_params = res.opt_SR_params ; % maximize R2 for the 35-sbj fit fprintf('\n Using optimized parameters: gamma=%.4f, lrate=%.4f \n',... SR_params(1), SR_params(3)) ; else % use the mean of the 35 cross-validation runs % See ??? for details... -- Taylor, please fill in SR_params = mean_CV_SR_params ; fprintf('\n Using fixed parameters: gamma=%.4f, lrate=%.4f \n',... SR_params(1), SR_params(3)) ; end % Run goalfun one last time, requesting full details in the export structure [cvRMSE,export] = LRaven1_R2_goalfun(SR_params,D,search_params) %#ok<NOPTS> % Store goalfun export in glbl structure glbl.descriptor = sprintf('Generated by LRaven1_R2_goalfun on %s',datestr(now,30)) ; glbl.SR_params = SR_params ; glbl.mean_SR_across_sbj = export.mean_SR_across_sbj ; glbl.std_SR_across_sbj = export.std_SR_across_sbj ; glbl.U = export.U ; glbl.Sigma = export.Sigma ; glbl.PC = export.PC ; glbl.projections = export.projections ; glbl.Borda_winners = export.Borda_winners ; glbl.R2_train = export.R2_train ; glbl.R2_test = export.R2_test ; glbl.RMSE_train = export.RMSE_train ; glbl.RMSE_test = export.RMSE_test ; glbl.Borda_gain = export.mean_R2_gain(:,glbl.Borda_winners) ; % Score targets target_subset = D.target(search_params.sbj_idx) ; mean_SR_across_sbj = export.mean_SR_across_sbj ; std_SR_across_sbj = export.std_SR_across_sbj ; PC = export.PC ; projections = export.projections(:,export.Borda_winners) ; % Global regression global_reg = regress_subset(target_subset,projections) ; beta = global_reg.beta ; R2_train1 = global_reg.R2 ; glbl.global_reg = global_reg ; b = global_reg.beta(1:N_comp) ; c = global_reg.beta(N_comp + 1) ; % Calculate global weights global_W1 = PC(:,glbl.Borda_winners) * b' ; % [N_vars x N_comp] * [N_comp x 1] = [N_vars x 1] glbl.global_weights1 = global_W1' ; % [1 x N_vars], without std_SR... global_W = global_W1 ./ std_SR_across_sbj' ; % [N_vars x 1] glbl.global_weights = global_W' ; % [1 x N_vars] %c = c - mean_SR_across_sbj * global_W ; c = c - mean_SR_across_sbj * global_W ; const_term = c ; % Define global M M = export.SR_by_sbj(search_params.sbj_idx,:) ; % Perform Prediction yhat = M * global_W + c ; glbl.predicted_targets = yhat' ; glbl.R2 = signed_R2(target_subset(:),glbl.predicted_targets(:)) ; glbl.RMSE = sqrt(mean((target_subset(:)-glbl.predicted_targets(:)).^2)) ; % Display glbl results in the published transcript glbl %#ok<NOPTS>
Using fixed parameters: gamma=0.2200, lrate=0.2300
cvRMSE =
2.5767
export =
search_params: [1x1 struct]
SR_params: [1x1 struct]
SR_by_sbj_by_trial: [35x28x100 double]
SR_by_sbj: [35x100 double]
mean_SR_across_sbj: [1x100 double]
std_SR_across_sbj: [1x100 double]
data: [35x100 double]
U: [35x35 double]
Sigma: [35x1 double]
PC: [100x35 double]
projections: [35x35 double]
mean_R2_gain: [1x35 double]
unanim_scores: [1.0417 0.9333 0.8417 0.7500 0.6667 0.5833 0.5000 0.4167 0.3333 0.2500 0.1667 0.0833]
ordered_scores: [1.0226 0.9369 0.8367 0.7532 0.6682 0.5843 0.5007 0.4173 0.3337 0.2503 0.1668 0.0834]
best_idx: [1 7 11 2 12 4 5 8 3 9 10 6]
Borda_winners: [1 7]
R2_set: 0.5137
R2_train: 0.5137
R2_test: 0.5137
RMSE_train: 2.5767
RMSE_test: 2.5767
glbl =
descriptor: 'Generated by LRaven1_R2_goalfun on 20110330T193609'
SR_params: [0.2200 0 0.2300]
mean_SR_across_sbj: [1x100 double]
std_SR_across_sbj: [1x100 double]
U: [35x35 double]
Sigma: [35x1 double]
PC: [100x35 double]
projections: [35x35 double]
Borda_winners: [1 7]
R2_train: 0.5137
R2_test: 0.5137
RMSE_train: 2.5767
RMSE_test: 2.5767
Borda_gain: [0.2708 0.2430]
global_reg: [1x1 struct]
global_weights1: [1x100 double]
global_weights: [1x100 double]
predicted_targets: [1x35 double]
R2: 0.5137
RMSE: 2.5767
090: Make quick and dirty plot of components, weights, and predictions
% Define plotting constants Tick=1:N_AOIs; TickLabel='1|2|3|4|5|6|7|8|9|R'; % Interpretation of the 10 AOIs % Set up colorbar poscol = [0 0 1]; negcol = [1 0 0]; pos_map = makecolormap(poscol,[1 1 1],5); neg_map = makecolormap([1 1 1],negcol,5); filler_map = ones(4,3); newmap = [pos_map; filler_map; neg_map]; % Make a plot with 3 comps, 1 global, 1 scatter of pred. vs observed % Plot global components clf ; %plot1 = figure('Position',[0 0 700 700],'PaperPositionMode','auto','Color','none') ; [rrr,ccc] = recommend_subplot_size(N_comp+1) ; for k = 1:search_params.N_comp subplot(2,2,k) ; PC(:,k) = glbl.PC(:,glbl.Borda_winners(k))*sign(glbl.global_reg.beta(k)) ; imagesc(reshape(PC(:,k),N_AOIs,N_AOIs),[-.18 .18]) ; colormap(newmap) %imagesc(reshape(PC(:,k),N_AOIs,N_AOIs),[-.21 .21]) ; title(sprintf('PC %d, R2g=%.3f, b=%.3f', ... glbl.Borda_winners(k),glbl.Borda_gain(k),abs(beta(k)))) ; set(gca,'XTick',Tick,'XTickLabel',TickLabel,'YTick',Tick,'YTickLabel',TickLabel); set(gca,'XTick',Tick+.50,'YTick',Tick+.50,'XTickLabel','','YTickLabel','') ; axis square ; grid on ; x = repmat(-.1,[1 10]) ; text(x,Tick,TickLabel) ; y = repmat(11,[1 10]) ; text(Tick-.15,y,TickLabel) ; end % %Plot global weights -- use weight1 which does NOT include std_SR_across_sbj subplot(2,2,N_comp+1) ; imagesc(reshape(glbl.global_weights1,N_AOIs,N_AOIs),[-.085,.085]) ; title('Weights') ; colormap(newmap) ; axis square ; %colorbar ; set(gca,'XTick',Tick,'XTickLabel',TickLabel,'YTick',Tick,'YTickLabel',TickLabel); set(gca,'XTick',Tick+.50,'YTick',Tick+.50,'XTickLabel','','YTickLabel','') ; axis square ; grid on ; x = repmat(-.1,[1 10]) ; text(x,Tick,TickLabel) ; y = repmat(11,[1 10]) ; text(Tick-.15,y,TickLabel) ; % Plot scatter of predicted and observed both sessions subplot(2,2,N_comp+2) plot(target_subset(:),glbl.predicted_targets(:),'o') ; %plot(target_subset(:),global_reg.yhat(sp2.sbj_idx),'o') ; title(sprintf('R2 = %.2f',round(glbl.R2*100)/100)) ; axis([0 29 0 29]) ; axis square ; grid on ; refline(1,0) ; xlabel('Observed score'); ylabel('Predicted score') ;
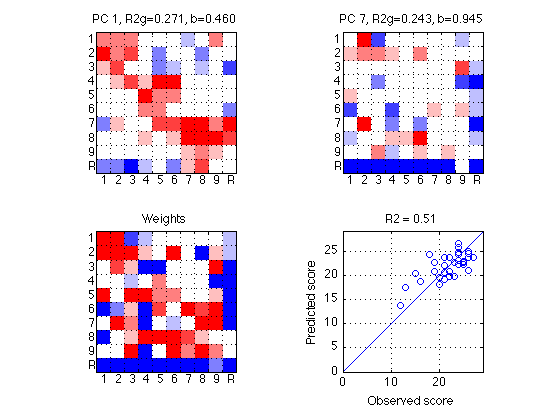
100: Generate publication-quality figures of components and weights
Generate each high resolution component separately and export of folder Use Omnigraffle for figure placement and final figure export
% Define plotting constants Tick=1:10; TickLabel='1|2|3|4|5|6|7|8|9|R'; % Define colorbar settings poscol = [0 0 1]; negcol = [1 0 0]; pos_map = makecolormap(poscol,[1 1 1],5); neg_map = makecolormap([1 1 1],negcol,5); filler_map = ones(4,3); newmap = [pos_map; filler_map; neg_map];
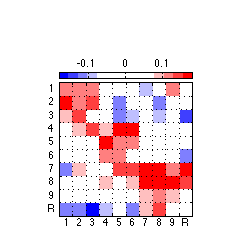
110: Generate and export high-quality systematicity component figure
clf ; %figure('Position',[0 0 175 175],'PaperPositionMode','auto','Color','none') ; % for image only figure('Position',[0 0 243 243],'PaperPositionMode','auto','Color','none') ; % for colorbar & image imagesc(reshape(PC(:,1),10,10),[-.18 .18]) ; colormap(newmap) ; c=colorbar('location','NorthOutside') ; % Position colorbar position and text x1=get(gca,'position'); x=get(c,'Position'); x(1)= .242 ; % move colorbar left/right %x(2)= .69 ; % move colorbar up/down x(2)= .68 ; % move colorbar up/down x(3)=.55 ; % length colorbar %x(4)=.04 ; % width colorbar x(4)=.025 ; % width colorbar set(c,'Position',x,'fontsize',8) ; % fontsize colorbar set(gca,'position',x1) ; % Adjust tick labels so they fall in the middle of matrix cells set(gca,'XTick',Tick+.50,'YTick',Tick+.50,'XTickLabel','','YTickLabel','') ; axis square ; grid on ; x = repmat(-.3,[1 10]) ; text(x,Tick,TickLabel,'fontsize',8) ; y = repmat(11,[1 10]) ; text(Tick-.227,y+.1,TickLabel,'fontsize',8) ; % Export 850 resolution png file to set folder % Uncomment this ... @@@ AAP 2011-03-30 %export_fig '~/work/RelExper/LRaven1/LRaven1_RESULTS/paper_figures/main_comp/syst_PC_fin.png' -png -r850 -a4

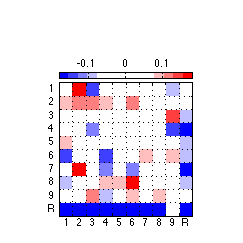
120: Generate and export high quality toggle component figure
clf ; %plot1 = figure('Position',[0 0 175 175],'PaperPositionMode','auto','Color','none') ; % for image only plot1 = figure('Position',[0 0 243 243],'PaperPositionMode','auto','Color','none') ; % for colorbar & image imagesc(reshape(PC(:,2),10,10),[-.18 .18]) ; colormap(newmap) ; c=colorbar('location','NorthOutside') ; % Position colorbar position and text x1=get(gca,'position'); x=get(c,'Position'); x(1)= .242 ; % move colorbar right x(2)= .68 ; % move colorbar up x(3)=.55 ; % length colorbar x(4)=.025 ; % width colorbar set(c,'Position',x,'fontsize',8) ; % fontsize colorbar set(gca,'position',x1) ; % Adjust tick labels so they fall in the middle of matrix cells set(gca,'XTick',Tick+.50,'YTick',Tick+.50,'XTickLabel','','YTickLabel','') ; axis square ; grid on ; x = repmat(-.3,[1 10]) ; text(x,Tick,TickLabel,'fontsize',8) ; y = repmat(11,[1 10]) ; text(Tick-.227,y+.1,TickLabel,'fontsize',8) ; % Export 850 resolution png file to set folder % Uncomment this ... @@@ AAP 2011-03-30 %export_fig '~/work/RelExper/LRaven1/LRaven1_RESULTS/paper_figures/main_comp/tog_PC_fin.png' -png -r850 -a4

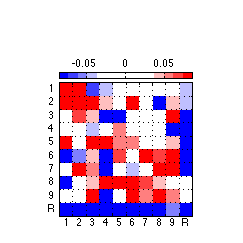
130: Generate and export high quality toggle weights figure
clf ; %figure('Position',[0 0 175 175],'PaperPositionMode','auto','Color','none') ; % for image only figure('Position',[0 0 243 243],'PaperPositionMode','auto','Color','none') ; % for colorbar & image imagesc(reshape(glbl.global_weights1,N_AOIs,N_AOIs),[-.085,.085]) ; colormap(newmap) ; c= colorbar('location','NorthOutside') ; % Position colorbar position and text x1=get(gca,'position'); x=get(c,'Position'); x(1)= .242 ; % move colorbar right x(2)= .68 ; % move colorbar up x(3)=.55 ; % length colorbar x(4)=.025 ; % width colorbar set(c,'Position',x,'fontsize',8) ; % fontsize colorbar set(gca,'position',x1) ; % Adjust tick labels so they fall in the middle of matrix cells set(gca,'XTick',Tick+.50,'YTick',Tick+.50,'XTickLabel','','YTickLabel','') ; axis square ; grid on ; x = repmat(-.3,[1 10]) ; text(x,Tick,TickLabel,'fontsize',8) ; y = repmat(11,[1 10]) ; text(Tick-.227,y+.1,TickLabel,'fontsize',8) ; % Export 850 resolution png file to set folder % Uncomment this ... @@@ AAP 2011-03-30 %export_fig '~/work/RelExper/LRaven1/LRaven1_RESULTS/paper_figures/main_comp/weights_fin.png' -png -r850 -a4

140:Generate and negative only weights
% Create positive only weight indices pos_idx = find(sign(glbl.global_weights1)==1) ; neg_idx = find(sign(glbl.global_weights1)==-1) ; % Generate zero vectors of 100 x 1 to store index values in pos_weights = zeros([100 1]) ; neg_weights = zeros([100 1]) ; % Positive weights pos_weights(pos_idx) = glbl.global_weights1(pos_idx) ; % Negative weights neg_weights(neg_idx) = glbl.global_weights1(neg_idx) ;
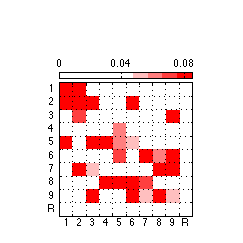
150: Generate and export high quality only positive weights figure
% Create a positive only colorbar negcol = [1 0 0]; neg_map = makecolormap([1 1 1],negcol,5); filler_map = ones(4,3); newmap = [filler_map; neg_map]; clf ; %figure('Position',[0 0 175 175],'PaperPositionMode','auto','Color','none') ; % for image only figure('Position',[0 0 243 243],'PaperPositionMode','auto','Color','none') ; % for colorbar & image imagesc(reshape(pos_weights,N_AOIs,N_AOIs),[0,.085]) ; colormap(newmap) ; c= colorbar('location','NorthOutside') ; % Position colorbar position and text x1=get(gca,'position'); x=get(c,'Position'); x(1)= .242 ; % move colorbar right x(2)= .68 ; % move colorbar up x(3)=.55 ; % length colorbar x(4)=.025 ; % width colorbar set(c,'Position',x,'fontsize',8,'XTick',0:.04:.08) ; % fontsize colorbar set(gca,'position',x1) ; % Adjust tick labels so they fall in the middle of matrix cells set(gca,'XTick',Tick+.50,'YTick',Tick+.50,'XTickLabel','','YTickLabel','') ; axis square ; grid on ; x = repmat(-.3,[1 10]) ; text(x,Tick,TickLabel,'fontsize',8) ; y = repmat(11,[1 10]) ; text(Tick-.227,y+.1,TickLabel,'fontsize',8) ; % Export 850 resolution png file to set folder % Uncomment this ... @@@ AAP 2011-03-30 %export_fig '~/work/RelExper/LRaven1/LRaven1_RESULTS/paper_figures/main_comp/pos_weights_fin.png' -png -r850 -a4

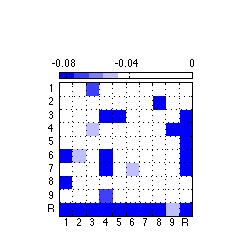
160: Generate and export high quality only negative weights figure
% Create a negative only colorbar poscol = [0 0 1]; pos_map = makecolormap(poscol,[1 1 1],5); filler_map = ones(4,3); newmap = [pos_map ; filler_map]; clf ; %figure('Position',[0 0 175 175],'PaperPositionMode','auto','Color','none') ; % for image only figure('Position',[0 0 243 243],'PaperPositionMode','auto','Color','none') ; % for colorbar & image imagesc(reshape(neg_weights,N_AOIs,N_AOIs),[-.085,0]) ; colormap(newmap) ; c= colorbar('location','NorthOutside') ; % Position colorbar position and text x1=get(gca,'position'); x=get(c,'Position'); x(1)= .242 ; % move colorbar right x(2)= .68 ; % move colorbar up x(3)=.55 ; % length colorbar x(4)=.025 ; % width colorbar set(c,'Position',x,'fontsize',8,'XTick',-.08:.04:.0) ; % fontsize colorbar set(gca,'position',x1) ; % Adjust tick labels so they fall in the middle of matrix cells set(gca,'XTick',Tick+.50,'YTick',Tick+.50,'XTickLabel','','YTickLabel','') ; axis square ; grid on ; x = repmat(-.3,[1 10]) ; text(x,Tick,TickLabel,'fontsize',8) ; y = repmat(11,[1 10]) ; text(Tick-.227,y+.1,TickLabel,'fontsize',8) ; % Export 850 resolution png file to set folder % Uncomment this ... @@@ AAP 2011-03-30 %export_fig '~/work/RelExper/LRaven1/LRaven1_RESULTS/paper_figures/main_comp/neg_weights_fin.png' -png -r850 -a4

999: Finish
close all %%%%% End of file LRaven1_gridcomp_optim.m